Nelder-Mead algorithm Plugin
Nelder-Mead Introduction
The Nelder-Mead plugin is used to fit an SBML model’s parameters to experimental data.
The plugin has numerous properties to allow the user full control over the internal fitting engine, as well as access to generated fitted data after a minimization session. In addition, various statistical properties, such as standardized residuals, Q-Q data, ChiSquare and reduced ChiSquare are made accessible to the user. The resulting parameter values also come with estimated confidence limits.
The current implementation is based on the Nelder-Mead C implementation by Michael F. Hutt 1.
1 An implementation of the Nelder-Mead simplex method. Copyright (c) 1997-2011 Michael F. Hutt
Nelder-Mead Plugin Properties
Available properties in the Nelder-Mead plugin are listed in the table below. The following properties are used internally by the fitting engine. They are pre-set with default values. Depending on the minimization problem at hand, they may need to be tweaked.
SBML: SBML document as a string. Model to be used in the fitting.
ExperimentalData: Input data.
FittedData: Output data.
InputParameterList: Parameters to fit.
OutputParameterList: List of fitted parameters.
ExperimentalDataSelectionList: Species selection list for experimental data.
FittedDataSelectionList: Selection list for model data.
Norm: Norm of fitting. An estimate of goodness of fit.
Norms: The norm is calculated throughout a fitting session. Each Norm value is stored in the Norms (read-only) property.
ConfidenceLimits: Confidence limits for each fitted parameter. The confidence limits are calculated at a 95% confidence level.
Hessian: Hessian matrix. The Hessian is calculated using approximation at a found parameter minimum.
CovarianceMatrix: Covariance matrix. Calculated as the inverse of the Hessian.
Residuals: Residuals data.
StandardizedResiduals: Standardized Residuals.
NormalProbabilityOfResiduals: Normal Probability of Residuals.
ChiSquare: The ChiSquare at the minimum.
ReducedChiSquare: The Reduced ChiSquare at the minimum.
StatusMessage: Message from the internal fitting engine, communicating the status of the obtained fit (Currently not used).
NrOfIter: Number of (internal outer loop) iterations.
NrOfFuncIter: Number of objective function iterations.
Epsilon: Convergence tolerance. Default is 1e-6.
Scale: Scaling of vertices. Default is 1.
MaxNrOfIterations: Maximum number of iterations. Default is 1000.
Alpha: Reflection coefficient. Default is 1.
Beta: Contraction coefficient. Default is 0.5.
Gamma: Expansion coefficient. Default is 1.
Nelder-Mead Plugin Events
The Nelder-Mead plugin uses all of the available plugin events, i.e. the PluginStarted, PluginProgress and the PluginFinished events. The available data variables for each event are internally treated as pass through variables, so any data, for any of the events, assigned prior to the plugin’s execute function (in the assignOn() family of functions), can be retrieved unmodified in the corresponding event function.
PluginStarted: Signals to application that the plugin has started applying noise on data. Both parameters are pass through parameters and are unused internally by the plugin.
PluginProgress: Communicates progress of noise generation. Both parameters are pass through parameters and are unused internally by the plugin.
PluginFinished: Signals to application that execution of the plugin has finished. Both parameters are pass through parameters and are unused internally by the plugin.
Python examples
The following Python script illustrates how the plugin can be used.
from rrplugins import *
# Load Plugins
chiPlugin = Plugin("tel_chisquare")
nm = Plugin("tel_nelder_mead")
modelPlugin = Plugin("tel_test_model")
addNoisePlugin = Plugin("tel_add_noise")
try:
#========== EVENT FUNCTION SETUP ===========================
def myEvent(dummy): #We are not capturing any data from the plugin, so just pass a dummy
print('Iteration, Norm = ' + `nm.getProperty("NrOfIter")` + ', ' + `nm.getProperty("Norm")`)
#Setup progress event function
progressEvent = NotifyEventEx(myEvent)
assignOnProgressEvent(nm.plugin, progressEvent)
#============================================================
#Create model data, with and without noise using the test_model plugin
modelPlugin.execute()
#Setup plugin properties.
nm.SBML = modelPlugin.Model
nm.ExperimentalData = modelPlugin.TestDataWithNoise
# Add the parameters that we're going to fit and an initial 'start' value
nm.setProperty("InputParameterList", ["k1", .3])
nm.setProperty("FittedDataSelectionList", "[S1] [S2]")
nm.setProperty("ExperimentalDataSelectionList", "[S1] [S2]")
# Start minimization
nm.execute()
print('Minimization finished. \n==== Result ====')
print('Hessian Matrix')
print(nm.getProperty("Hessian"))
print('Covariance Matrix')
print(nm.getProperty("CovarianceMatrix"))
print('ChiSquare = ' + `nm.getProperty("ChiSquare")`)
print('Reduced ChiSquare = ' + `nm.getProperty("ReducedChiSquare")`)
#This is a list of parameters
parameters = tpc.getPluginProperty (nm.plugin, "OutputParameterList")
confLimits = tpc.getPluginProperty (nm.plugin, "ConfidenceLimits")
#Iterate trough list of parameters and confidence limits
para = getFirstProperty(parameters)
limit = getFirstProperty(confLimits)
while para and limit:
print(getPropertyName(para) + ' = ' + `getPropertyValue(para)` + ' +/- ' + `getPropertyValue(limit)`)
para = getNextProperty(parameters)
limit = getNextProperty(confLimits)
# Get the fitted and residual data
fittedData = nm.getProperty ("FittedData").toNumpy
residuals = nm.getProperty ("Residuals").toNumpy
# Get the experimental data as a numpy array
experimentalData = modelPlugin.TestDataWithNoise.toNumpy
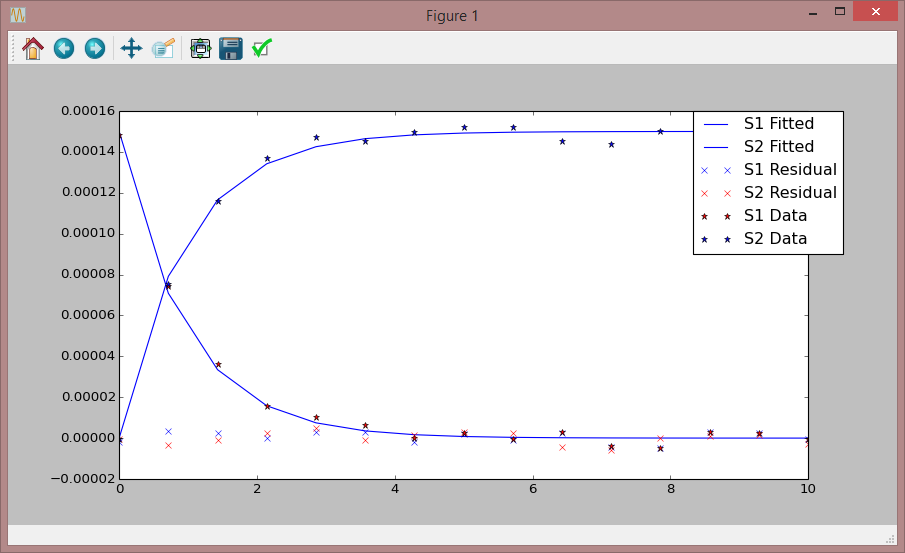
telplugins.plot(fittedData [:,[0,1]], "blue", "-", "", "S1 Fitted")
telplugins.plot(fittedData [:,[0,2]], "blue", "-", "", "S2 Fitted")
telplugins.plot(residuals [:,[0,1]], "blue", "None", "x", "S1 Residual")
telplugins.plot(residuals [:,[0,2]], "red", "None", "x", "S2 Residual")
telplugins.plot(experimentalData [:,[0,1]], "red", "", "*", "S1 Data")
telplugins.plot(experimentalData [:,[0,2]], "blue", "", "*", "S2 Data")
telplugins.plt.show()
#Finally, view the manual and version
nm.viewManual()
print('Plugin version: ' + `nm.getVersion()`)
except Exception as e:
print('Problem.. ' + `e`)