The Main RoadRunner Class
- class RoadRunner
The main RoadRunner class. Its objects, i.e.
rr = RoadRunner()perform the libRoaRunner functions, i.e.rr.simulate().
- RoadRunner.__init__(uriOrSBML='', options=None)
Creates a new RoadRunner object. If the first argument is specified, it should be a string containing either the contents of an SBML document, or a formatted URI specifying the path or location of a SBML document.
If options is given, it should be a LoadSBMLOptions object.
If no arguments are given, a document may be loaded at any future time using the load method.
- Parameters:
uriOrSBML – a URI, local path or SBML document contents.
options – (LoadSBMLOptions) an options object specifying how the SBML document should be loaded
- RoadRunner.load(uriOrDocument)
Loads an SBML document, given a string for file path, URI, or contents.
This method also accepts HTTP URI for remote files, however this feature is currently limited to the Mac version, plan on enabling HTTP loading of SBML documents on Windows and Linux shortly.
Some examples of loading files on Mac or Linux:
>>> rr.load("myfile.xml") # load a file from the current directory >>> rr.load("/Users/Fred/myfile.xml") # absolute path >>> rr.load("http://sbml.org/example_system.xml") # remote file
Or on Windows:
>>> rr.load("myfile.xml") # load a file from the current directory >>> rr.load("file://localhost/c:/Users/Fred/myfile.xml") # using a URI
One may also load the contents of a document:
>>> myfile = open("myfile.xml, "r") >>> contents = file.read() >>> rr.load(contents)
Loading in a raw SBML string is also possible:
>>> sbmlstr = rr.getCurrentSBML() # Or any other properly formatted SBML string block >>> rr.load(sbmlstr)
In future version, we will also support loading directly from a libSBML Document object.
- Parameters:
uriOrDocument – A string which may be a local path, URI or contents of an SBML document.
- RoadRunner.saveState(document, option='b')
Saves the current state of the RoadRunner instance, e.g. integrator, steady state solver, simulation results, given a string for file path. If no option is given or the option is ‘b’, the state will be stored in a binary format which can be quickly reloaded for later simulation. This binary format is platform specific. It the option is ‘r’, the state will be stored in a human-readable format which can be used for debugging, but cannot be reloaded later.
Some examples of saving binary files on Mac or Linux:
>>> rr.saveState("current_state.txt") # save the state to a file from the current directory >>> rr.saveState("/Users/Fred/current_state.txt") # absolute path
Or on Windows:
>>> rr.saveState("current_state.txt") # save the state to a file from the current directory >>> rr.saveState("file://localhost/c:/Users/Fred/current_state.txt") # using a URI
One may also save in a human-readable format:
>>> rr.saveState("current_state.txt", 'r')
- Parameters:
document – The file path where the current state will be stored
option – an option object specifying how the state should be saved ‘b’ - binary (default) ‘r’ - human-readable
- RoadRunner.loadState(document)
Loads the saved state of a RoadRunner instance, e.g. integrator, steady state solver, simulation results, given a string for file path. All simulation calls after this function will start from the resumed state.
Some examples of reloading binary files on Mac or Linux:
>>> rr.loadState("current_state.txt") # load the state from a file from the current directory >>> rr.loadState("/Users/Fred/current_state.txt") # absolute path
Or on Windows:
>>> rr.loadState("current_state.txt") # load the state from a file from the current directory >>> rr.loadState("file://localhost/c:/Users/Fred/current_state.txt") # using a URI
- Parameters:
document – The file path where the state of simulation will be loaded from
- RoadRunner.getCompiler()
Returns the JIT
Compilerobject currently being used. This object provides various information about the current processor and system.
- RoadRunner.getConfigurationXML()
Recurse through all of the child configurable objects that this class ones and build an assemble all of their configuration parameters into a single xml document which is returned as a string.
The value of this result depends on what child objects are presently loaded.
- RoadRunner.getExtendedVersionInfo()
Returns
getVersionStr()as well as info about dependent libs versions.
- RoadRunner.getInfo()
Returns info about the current state of the object.
- Return type:
str
- RoadRunner.getInstanceCount()
Returns number of currently running RoadRunner instances.
- RoadRunner.getInstanceID()
Returns the instance id when there are multiple instances of RoadRunner.
- RoadRunner.getIntegrator()
Returns the solver instance. See :cpp:class:
rr::roadrunner::Solver. For more information on the possible settings, see Solver Class.
- RoadRunner.getIntegratorByName(name)
Returns the solver instance by given name. See
rr::roadrunner::Solver. For more information on the possible settings, see Solver Class.- Parameters:
name (str) – Name of the integrator
- RoadRunner.getAvailableIntegrators()
Returns a list of names of available integrators.
- RoadRunner.getExistingIntegratorNames()
Returns a list of names of all integrators.
- RoadRunner.getParamPromotedSBML(*args)
Takes an SBML document (in textual form) and changes all of the local parameters to be global parameters.
- Parameters:
SBML (str) – the contents of an SBML document
- Return type:
str
- RoadRunner.getCurrentSBML()
Returns the SBML with the current model parameters. This is different than
getSBML()which returns the original SBML. This may optionally up or down-convert the document to a different version, if the level and version arguments are non-zero. If both arguments are zero, then the document is left alone and the original version is returned.- Return type:
str
- RoadRunner.getSBML()
Returns the original SBML model that was loaded into roadrunner. If the model is edited by methods in editing section, it will return the most updated model with the initial model parameters.
- Return type:
str
- RoadRunner.setIntegrator(name)
Sets specific integrator. For more information on the possible settings, see Solver Class.
- Parameters:
name (str) – name of the integrator.
- RoadRunner.setIntegratorSetting(name, key, value)
Sets settings for a specific integrator. See
roadrunner::Solver. For more information on the possible settings, see Solver Class.- Parameters:
name (str) – name of the integrator.
key (str) – name of the setting.
value (const) – value of the setting.
Selections
- RoadRunner.getIds()
Return a list of selection ids that this object can select on.
- Return type:
list
- RoadRunner.[element_id]
Access the current value of a model element. All model elements with mathematical meaning (species, compartment, or parameters) can be accessed and set using this method. >>> rr.S1 6.3 >>> rr.S1 = 2.9 >>> rr.S1 2.9
- RoadRunner.getValue(sel)
Returns the value for a given selection. For more information on accepted selection types see Selecting Values.
- Parameters:
sel (str or SelectionRecord) – a selection that is either a string or a SelectionRecord that was obtained from createSelection
- RoadRunner.setValue(sel, value)
Sets the value for a given selection. For more information on accepted selection types see Selecting Values.
- Parameters:
sel (str) – a selection that is either a string or a SelectionRecord that was obtained from createSelection
value (double) – the value of the selection to be set
- RoadRunner.setValues(keysOrDict, values=None)
Sets a number of values in the roadrunner object all at once. Use either with the first argument defined as a dictionary, or with both arguments defined, with the first as the keys and the second as the values. see Selecting Values.
>>> parameters = {'a': 7, 'b': 8} >>> rr.setValues(parameters) >>> rr.setValues(parameters.keys(), parameters.values())
- Parameters:
keysOrDict (list<str> or dict<str, double>) – either a list of id strings to set, or a dictionary with string keys and numerical values.
value (double) – the list of values to use. Must be identical in length to ‘keysOrDict’, and keysOrDict must not be a dictionary.
- RoadRunner.getSelectedValues()
returns the values of the current timecourse selections for the current state of the model
- Return type:
numpy.ndarray
- RoadRunner.timeCourseSelections
Get or set the list of current selections used for the time course simulation result columns. For more information on accepted selection types, see Selecting Values.
>>> rr.timeCourseSelections = ['time', 'S1', 'S2'] >>> rr.timeCourseSelections ['time', 'S1', 'S2']
- RoadRunner.createSelection(sel)
Create a new selection based on a selection string
- Return type:
- RoadRunner.resetSelectionLists()
Resets time course and steady state selection lists to defaults
Model Access
- RoadRunner.getValue(eid)
The current value of model elements can be obtained or set by using the ID of the element. This works for all model elements with mathematical meaning (species, compartments, or parameters) >>> rr.S1 6.3 >>> rr.S1 = 2.9 >>> rr.S1 2.9
- RoadRunner.isModelLoaded()
Return True if model was loaded; False otherwise
- RoadRunner.model
Get the currently loaded model. The model object contains the entire state of the SBML model.
- RoadRunner.getModel()
Function form of the RoadRunner.model property, identical to model.
- RoadRunner.clearModel()
Clears the currently loaded model and all associated memory. Returns True if memory was freed, False if no model was loaded in the first place.
>>> rr.isModelLoaded() True >>> rr.clearModel() >>> rr.isModelLoaded() False
- RoadRunner.oneStep(startTime, stepSize)
Carry out a one step integration of the model. The method takes two arguments, the current time and the step size to use in the integration. Returs the new time which will be currentTime + StepSize:
newTime = rr.oneStep (10, 0.5)
- RoadRunner.internalOneStep(startTime, stepSize, reset)
Carry out a single internal solver integration step. The method takes three arguments, the current time and the step size to use in the integration and reset. Reset defaults to true, set to false to stop integrator instance from restarting. Returns the end time.
- RoadRunner.reset()
Resets time, all floating species, and rates to their initial values. Does NOT reset changed global parameters.
- RoadRunner.resetAll()
Resets time, all floating species, and rates to their CURRENT initial values. Also resets all global parameters back to the values they had when the model was first loaded. “Current” initial values are set by using
rr.setValue('init(S1)', 5)which sets a species named S1 to have current initial value of 5. Note it is NOT the initial values of when the model was first loaded in.
- RoadRunner.resetParameter()
Resets only global parameters to their CURRENT initial values.
- RoadRunner.resetToOrigin()
Resets the model back to the state it was when FIRST loaded. The scope of this reset includes all initial values and parameters (everything).
- RoadRunner.setConfigurationXML(*args)
Given a xml document, which should have been returned from getConfigurationXML, this method recurses though all the child configurable elements and sets their configuration to the values specified in the document.
- Parameters:
xml (str) – the contents of an xml document.
- RoadRunner.conservedMoietyAnalysis
Enables / Disables conserved moiety analysis (boolean).
If this is enabled, the SBML document (either current, or one about to be loaded) is converted using the ConservedMoietyConverter. All of the linearly dependent species are replaced with assignment rules and a new set of conserved moiety parameters are introduced.
To enable, type:
>>> rr.conservedMoietyAnalysis = True
Model Editing
Easy edit to the model without modifying and reloading sbml files.
- RoadRunner.addSpeciesConcentration(sid, compartment, initValue, substanceUnits, forceRegenerate)
Add a species to the current model. Note that the species to be added must have an ID that did not existed in the model. The given compartment must also exist in the model.
Default substanceUnits is “concentration”, which will set initValue as initial concentration of the new species. Other substanceUnits will set initValue as initial amount of the new species.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.addSpeciesConcentration("s1", "compartment", 0.1, False) # it will not regenerate the model, nothing actually happened >>> rr.addSpeciesConcentration("s2", "compartment", 0.1, True) # new model is generated and saved
- Parameters:
sid (str) – the ID of the species to be added
compartment (str) – the compartment of the species to be added
initValue (double) – the initial amount or concentration of the species to be added
substanceUnits (str) – the substance unit of the species to be added
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.removeSpecies(sid, forceRegenerate)
Remove a species from the current model. Note that the given species must exist in the current model.
All reactions related to this species(as reactants, products or modifiers or used in stoichiometry) will be removed. Kinetic law used this species in the math formula will be unset. All function definitions, constraints, initial assignments and rules related to this species (as variables or used in math formula) will be removed. All events used this speices in trigger formula will be removed. Priority and delay used this sepcies in the math formula will be unset. All event assignment related to this species(as variables or used in math formula) will be removed.
If any global parameters become uninitialized during this process, i.e, has no initial assignment or assignment rule, they will be removed recursively following the rules in removeParameter().
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.removeSpecies("s1", false) # it will not regenerate the model, nothing actually happened >>> rr.removeSpecies("s2", true) # new model is generated and saved
- Parameters:
sid (str) – the ID of the species to be removed
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addReaction(rid, reactants, products, kineticLaw, forceRegenerate)
Add a reaction to the current model by passing its info as parameters. Note that the reaction to be added must have an ID that did not existed in the model.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.addReaction("r1", ["s1"], ["s2"], "s1 * k1", False) # it will not regenerate the model, nothing actually happened >>> rr.addReaction("r2", ["s2"], ["s1"], "s2 * k1", True) # new model is generated and saved
- Parameters:
rid (str) – the ID of the reaction to be added
reactants (list) – the list of reactants ID of reaction to be added
products (list) – the list of products ID of reaction to be added
kineticLaw (str) – the kinetic formular of reaction to be added
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addReaction(sbmlRep, forceRegenerate)
Add a reaction to the current model by passing a sbml repesentation as parameter. Note that the reaction to be added must have an ID that did not existed in the model.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
- Parameters:
sbmlRep (str) – the SBML representation (i.e. a reaction tag) describing the reaction to be added
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.removeReaction(rid, forceRegenerate)
Remove a reaction from the current model. Note that the given reaction must exist in the current model.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.removeReaction("r1", False) # it will not regenerate the model, nothing actually happened >>> rr.removeReaction("r2", True) # new model is generated and saved
- Parameters:
rid (str) – the ID of the reaction to be removed
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addParameter(pid, value, forceRegenerate)
Add a parameter to the current model. Note that the parameter to be added must have an ID that did not existed in the model.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.addParameter("p1", 0.1, False) # it will not regenerate the model, nothing actually happened >>> rr.addParameter("p2", 0.1, True) # new model is generated and saved
- Parameters:
pid (str) – the ID of the parameter to be added
value (double) – the initial value of the parameter to be added
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.removeParameter(pid, forceRegenerate)
Remove a parameter from the current model. Note that the given parameter must exist in the current model.
All reactions related to this parameter(used in stoichiometry) will be removed. Kinetic law used this parameter in the math formula will be unset. All function definitions, constraints, initial assignments and rules related to this parameter (as variables or used in math formula) will be removed. All events used this parameter in trigger formula will be removed. Priority and delay used this parameter in the math formula will be unset. All event assignment related to this parameter(as variables or used in math formula) will be removed.
If any global parameters become uninitialized during this process, i.e, has no initial assignment or assignment rule, they will be removed recursively following the above rules.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.removeParameter("p1", False) # it will not regenerate the model, nothing actually happened >>> rr.removeParameter("p2", True) # new model is generated and saved
- Parameters:
pid (str) – the ID of the parameter to be removed
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addCompartment(cid, initVolume, forceRegenerate)
Add a compartment to the current model. Note that the compartment to be added must have an ID that did not existed in the model.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.addCompartment("c1", 0.1, False) # it will not regenerate the model, nothing actually happened >>> rr.addCompartment("c2", 0.1, True) # new model is generated and saved
- Parameters:
cid (str) – the ID of the compartment to be added
initVolume (double) – the initial volume of the compartment to be added
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.removeCompartment(cid, forceRegenerate)
Remove a compartment from the current model. Note that the given compartment must exist in the current model.
All reactions related to this compartment(used in stoichiometry) will be removed. Kinetic law used this compartment in the math formula will be unset. All function definitions, constraints, initial assignments and rules related to this compartment (as variables or used in math formula) will be removed. All events used this compartment in trigger formula will be removed. Priority and delay used this compartment in the math formula will be unset. All event assignment related to this compartment(as variables or used in math formula) will be removed.
If any global parameters become uninitialized during this process, i.e, has no initial assignment or assignment rule, they will be removed recursively following the rules in removeParameter().
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.removeCompartment("c1", False) # it will not regenerate the model, nothing actually happened >>> rr.removeCompartment("c2", True) # new model is generated and saved
- Parameters:
cid (str) – the ID of the compartment to be removed
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.setKineticLaw(rid, kineticLaw, forceRegenerate)
Set hte kinetic law for an existing reaction in the current model. Note that given reaction must exist in the model.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.setKineticLaw("r1", "s1 * k1", False) # it will not regenerate the model, nothing actually happened >>> rr.setKineticLaw("r2", "s2 * k1", True) # new model is generated and saved
- Parameters:
rid (str) – the ID of the reaction to be modified
kineticLaw (str) – the kinetic formular of reaction to be set
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addAssignmentRule(vid, formula, forceRegenerate)
Add an assignment rule for a variable to the current model.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.addAssignmentRule("s1", "s1 * k1", False) # it will not regenerate the model, nothing actually happened >>> rr.addAssignmentRule("s2", "s2 * k1", True) # new model is generated and saved
- Parameters:
vid (str) – the ID of the variable that the new rule assigns formula to
formula (str) – the math formula of assignment rule to be added
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addRateRule(vid, formula, forceRegenerate)
Add a rate rule for a variable to the current model.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.addRateRule("s1", "k1", False) # it will not regenerate the model, nothing actually happened >>> rr.addRateRule("s2", "k1", True) # new model is generated and saved
- Parameters:
vid (str) – the ID of the variable that the new rule assigns formula to
formula (str) – the math formula of rate rule to be added
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.removeRules(vid, forceRegenerate)
Remove all rules for a variable from the current model, including assignment and rate rules. Note that the given variable must have at least one rule in the current model.
If any global parameters become uninitialized during this process, i.e, has no initial assignment or assignment rule, they will be removed recursively following the rules in removeParameter().
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.removeRules("s1", False) # it will not regenerate the model, nothing actually happened >>> rr.removeRules("s2", True) # new model is generated and saved
- Parameters:
vid (str) – the ID of the variables that rules assign formula to
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addEvent(eid, useValuesFromTriggerTime, trigger, forceRegenerate)
Add an event to the current model. Note that the event to be added must have an ID that did not existed in the model.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.addEvent("e1", False, "s1 > 0", False) # it will not regenerate the model, nothing actually happened >>> rr.addEvent("e2", False, "s2 == s1", True) # new model is generated and saved
- Parameters:
eid (str) – the ID of the event to be added
useValuesFromTriggerTime (bool) – indicate the moment at which the event’s assignments are to be evaluated
trigger (str) – the math formula of event trigger
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addTrigger(eid, trigger, forceRegenerate)
Add trigger to an existing event in the model. Note that the given event must exist in the current model. If the given event already has a trigger object, the given trigger will replace the old trigger of the event.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.addTrigger("e1", "s1 > 0", False) # it will not regenerate the model, nothing actually happened >>> rr.addTrigger("e2", "s2 == s1", True) # new model is generated and saved
- Parameters:
eid (str) – the ID of the event to add the trigger to
trigger (str) – the math formula of event trigger
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addPriority(eid, priority, forceRegenerate)
Add priority to an existing event in the model. Note that the given event must exist in the current model. If the given event already has a priority object, the given priority will replace the old priority of the event.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
- Parameters:
eid (str) – the ID of the event to add the priority to
priority (str) – the math formula of event priority
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addDelay(eid, delay, forceRegenerate)
Add delay to an existing event in the model. Note that the given event must exist in the current model. If the given event already has a delay object, the given delay will replace the old delay of the event.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
- Parameters:
eid (str) – the ID of the event to add the delay to
delay (str) – the math formula of event delay
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.addEventAssignment(eid, vid, formula, forceRegenerate)
Add an event assignment to an existing event in the model. Note that the given event must exist in the current model.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.addEventAssignment("e1", "s1", "k1", False) # it will not regenerate the model, nothing actually happened >>> rr.addEventAssignment("e2", "s2", "s1", True) # new model is generated and saved
- Parameters:
eid (str) – the ID of the event to add the event assignment to
vid (str) – the ID of the variables that assignment assigns formula to
formula (str) – the math formula of event assignment
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.removeEventAssignment(eid, vid, forceRegenerate)
Add all event assignments for a variable from an existing event in the model. Note that the given event must exist in the current model and given variable must have an event assignment in the given event.
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.removeEventAssignment("e1", "s1", False) # it will not regenerate the model, nothing actually happened >>> rr.removeEventAssignment("e2", "s2", True) # new model is generated and saved
- Parameters:
eid (str) – the ID of the event
vid (str) – the ID of the variables of the event assignments to be removed
forceRegenerate (bool) – indicate whether the new model is regenerated after this function call
- RoadRunner.removeEvent(eid, forceRegenerate)
- :module: RoadRunner
Add an event from the current model. Note that the given event must exist in the current model.
If any global parameters become uninitialized during this process, i.e, has no initial assignment or assignment rule, they will be removed recursively following the rules in removeParameter().
forceRegenerate is a boolean value that indicates whether the new model will be regenerated. Its default value is true, which means to regenerate model every time after this function is called. Note that regenerating model is time-consuming. To save time for editing model for multiple times, one could set this flag to false excepting for the last call, so that Roadrunner will only regenerate the model once after all editings are completed.
For example,
>>> rr.removeEvent("e1", False) # it will not regenerate the model, nothing actually happened >>> rr.removeEvent("e2", True) # new model is generated and saved
- param str eid:
the ID of the event to be removed
- param bool forceRegenerate:
indicate whether the new model is regenerated after this function call.
- regenerateModel()
- :module RoadRunner
Call this method to jit compile any model you’ve constructed using the modeling editing API. This will make the model ready for simulation.
For example:
>>>> rr.addCompartment (‘c1’, 0.1) >>>> rr.addSpeciesConcentration (‘s1’, ‘c1’, 1.5, False, False) >>>> rr.addSpeciesConcentration (‘s2’, ‘c1’, 0.0, False, False) >>>> rr.addParameter(‘k1’, 0.2) >>>> rr.addReaction (‘r1’, [‘s1’], [‘s2’], ‘s1*k1’) >>>> rr.regenerateModel() >>>> rr.simulate()
Simulation
Fast and easy time series simulations is one of the main objectives of the RoadRunner project.
All simulation related tasks can be accomplished with the single simulate method.
- RoadRunner.simulate(*args, **kwargs)
Simulate current SBML model.
simulate accepts up to five positional arguments.
The first five (optional) arguments are treated as:
1: Start Time, if this is a number.
2: End Time, if this is a number.
3: Number of points, if this is a number.
4: List of Selections. A list of variables to include in the output, e.g.
['time','A']for a model with speciesA. More below.5: output file path. The file to which simulation results will be written. If this is specified and nonempty, simulation output will be written to output_file every Config::K_ROWS_PER_WRITE generated. Note that simulate() will not return the result matrix if it is writing to output_file. It will also not keep any simulation data, so in that case one should not call
rr.plot()without arguments. This should be specified when one cannot, or does not want to, keep the entire result matrix in memory.All five of the positional arguments are optional. If any of the positional arguments are a list of string instead of a number, then they are interpreted as a list of selections.
By default, the gillespie algorithm output uses a variable step size. However, if both the end time and the number of steps is defined, the output is constrained to a grid instead. If just the end time is defined, a variable number of steps will be taken until that end time is reached, or until the maximum number of output rows is reached. If just the number of steps is defined, that many variable steps will be taken.
The maximum number of output rows is set to 100,000 by default, though this is ignored if the variable step size is set to false, or if the output is being written directly to a file. It can be changed by with the argument ‘max_output_rows’.
There are a number of ways to call simulate.
- 1: With no arguments. In this case, the current set of options from the previous
simulatecall will be used. If this is the first timesimulateis called, then a default set of values is used. The default set of values are (start = 0, end = 5).
2: With up to five positions arguments, described above.
Finally, you can pass steps keyword argument instead of points.
steps (Optional) Number of steps at which the output is sampled where the samples are evenly spaced. Steps = points-1. Steps and points may not both be specified.
- Returns:
a numpy array with each selected output time series being a column vector, and the 0’th column is the simulation time; or if output_file is specified and nonempty, a message string
- Return type:
numpy.ndarray
- RoadRunner.gillespie(start, end, steps)
Run a Gillespie stochastic simulation.
Use
RoadRunner.reset()to reset the model each time.- Parameters:
start – start time
end – end time
steps – number of steps
- Returns:
a numpy array with each selected output time series being a column vector, and the 0’th column is the simulation time.
- Return type:
numpy.ndarray
Examples:
Simulate from time zero to 40 time units
>>> result = rr.gillespie (0, 40)
Simulate on a grid with 10 points from start 0 to end time 40
>>> result = rr.gillespie (0, 40, 10)
Simulate from time zero to 40 time units using the given selection list
>>> result = rr.gillespie (0, 40, [‘time’, ‘S1’])
Simulate from time zero to 40 time units, on a grid with 20 points using the given selection list
>>> result = rr.gillespie (0, 40, 20, [‘time’, ‘S1’])
- RoadRunner.plot(result=None, loc='upper left', show=True)
Plot results from a simulation carried out by the simulate or gillespie functions.
To plot data currently held by roadrunner that was generated in the last simulation, use:
>>> rr.plot()
If you are using Tellurium, see tellurium.ExtendedRoadRunner.plot which supports extra arguements.
- Parameters:
result (numpy.ndarray) – Data returned from a simulate or gillespie call
loc (str) – string representing the location of legend i.e. “upper right”
- Roadrunner.getSimulationData()
Returns the array of simulated data. When simulation has not been run, the function will return an empty array.
Steady State
- class RoadRunner.SteadyStateSolver
RoadRunner.SteadyStateSolver class.
- RoadRunner.steadyStateSelections
A list of SelectionRecords which determine what values are used for a steady state calculation. This list may be set by assigning a list of valid selection symbols:
>>> rr.steadyStateSelections = ['S1', '[S2]', 'P1'] >>> rr.steadyStateSelections ['S1', '[S2]', 'P1']
- RoadRunner.steadyState()
Attempts to evaluate the steady state for the model. The method returns a value that indicates how close the solution is to the steady state. The smaller the value the better. Values less than 1E-6 usually indicate a steady state has been found. If necessary the method can be called a second time to improve the solution.
- Returns:
the sum of squares of the steady state solution.
- Return type:
double
- RoadRunner.getSteadyStateValues()
Performs a steady state calculation (evolves the system to a steady state), then calculates and returns the set of values specified by the steady state selections.
- Returns:
a numpy array corresponding to the values specified by steadyStateSelections
- Return type:
numpy.ndarray
- RoadRunner.getSteadyStateValuesNamedArray()
Performs a steady state calculation (evolves the system to a steady state), then calculates and returns the set of values specified by the steady state selections with all necessary labels.
- Returns:
a NamedArray corresponding to the values specified by steadyStateSelections
- Return type:
NamedArray
- RoadRunner.getSteadyStateSolver()
Returns the steady state solver which is currently being used.
- RoadRunner.steadyStateSolverExists(name)
Checks whether a steady state solver exists.
- Parameters:
name (str) – name of a steady state solver
Metabolic control analysis
In the special case when an SBML model is a purely reaction kinetics model – no rate rules, no assignment rules for chemical species, and time invariant stoichiometry, specialized analysis methods related to metabolic control analysis are applicable. These methods are described in this section.
- RoadRunner.getCC(variable, parameter)
Returns a scaled control coefficient with respect to a global parameter.
For example:
rr.getCC ('J1', 'Vmax') rr.getCC ('S1', 'Xo') rr.getCC ('S2', 'Km')
The first returns a flux control coefficient with respect to flux J1. The second and third return concentration control coefficients with respect to species S1 and S2.
- Parameters:
variable – The id of a dependent variable of the coefficient, for example a reaction or species concentration.
parameter – The id of the independent parameter, for example a kinetic constant or boundary species
- Returns:
the value of the scaled control coefficient.
- Return type:
double
- RoadRunner.getuCC(variableId, parameterId)
Get unscaled control coefficient with respect to a global parameter.
- Parameters:
variableId – The id of a dependent variable of the coefficient, for example a reaction or species concentration.
parameterId – The id of the independent parameter, for example a kinetic constant or boundary species
- Returns:
the value of the unscaled control coefficient.
- Return type:
double
- RoadRunner.getEE(reactionId, parameterId, steadyState=True)
Retrieve a single elasticity coefficient with respect to a global parameter.
For example:
x = rr.getEE ('J1', 'Vmax')
calculates elasticity coefficient of reaction ‘J1’ with restpect to parameter ‘Vmax’.
- Parameters:
variable (str) – A reaction Id
parameter (str) – The independent parameter, for example a kinetic constant, floating or boundary species
steadyState (Boolean) – should the steady state value be computed.
- RoadRunner.getuEE(reactionId, parameterId)
Get unscaled elasticity coefficient with respect to a global parameter or species.
- RoadRunner.getEigenValueIds()
Returns a list of selection symbols for the eigenvalues of the floating species. The eigen value selection symbol is
eigen(XX), whereXXis the floating species name.
- RoadRunner.getFullEigenValues()
Calculates the eigen values of the Full Jacobian as a real matrix, first column real part, second column imaginary part.
Note, only valid for pure reaction kinetics models (no rate rules, no floating species rules and time invariant stoichiometry).
- Return type:
numpy.ndarray
- RoadRunner.getReducedEigenValues()
Calculates the eigen values of the Reduced Jacobian as a real matrix, first column real part, second column imaginary part.
Only valid if moiety conversion is enabled.
Note, only valid for pure reaction kinetics models (no rate rules, no floating species rules and time invariant stoichiometry).
- Return type:
numpy.ndarray
- RoadRunner.getFullJacobian()
Compute the full Jacobian at the current operating point.
This is the Jacobian of ONLY the floating species.
- RoadRunner.getReducedJacobian()
Returns the reduced Jacobian for the independent species. This matrix will be non-singular for models that include moiety-conserved cycles.
- Return type:
numpy.ndarray
- RoadRunner.getScaledConcentrationControlCoefficientMatrix()
Returns the m by n matrix of scaled concentration control coefficients where m is the number of floating species and n the number of reactions.
- Return type:
numpy.ndarray
- RoadRunner.getScaledFloatingSpeciesElasticity(reactionId, speciesId)
Returns the scaled elasticity for a given reaction and given species.
- Parameters:
reactionId (str) – the SBML id of a reaction.
speciesId (str) – the SBML id of a species.
- Return type:
double
- RoadRunner.getUnscaledParameterElasticity(reactionId, parameterId)
Returns the unscaled elasticity for a named reaction with respect to a named parameter
- Parameters:
reactionId (str) – the SBML id of a reaction.
parameterId (str) – the SBML id of a parameter.
- Return type:
double
- RoadRunner.getUnscaledConcentrationControlCoefficientMatrix()
Returns the unscaled concentration control coefficient matrix.
- RoadRunner.getUnscaledElasticityMatrix()
Returns the unscaled species elasticity matrix at the current operating point.
- RoadRunner.getUnscaledFluxControlCoefficientMatrix()
Returns the unscaled flux control coefficient matrix.
- RoadRunner.getUnscaledSpeciesElasticity(reactionIndx, speciesIndx)
Get a single species elasticity value.
- Parameters:
reactionIndx (int) – index of reaction
speciesIndx (int) – index of species.
- RoadRunner.getScaledFluxControlCoefficientMatrix()
Returns the n by n matrix of scaled flux control coefficients where n is the number of reactions.
- Return type:
numpy.ndarray
- RoadRunner.getScaledElasticityMatrix()
Returns the scaled elasticity matrix at the current operating point.
- Return type:
numpy.ndarray
- RoadRunner.setDiffStepSize(val)
Sets the differential step size used in routines such as
getCC().- Parameters:
val – differential step size
Stoichiometric Analysis
- RoadRunner.getFullStoichiometryMatrix()
Get the stoichiometry matrix that coresponds to the full model, even it it was converted via conservation conversion.
- RoadRunner.getReducedStoichiometryMatrix()
Get the reduced stochiometry matrix. If conservation conversion is enabled, this is the matrix that coresponds to the independent species.
A synonym for getNrMatrix().
- RoadRunner.getConservationMatrix()
Returns a conservation matrix
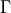 which is a
which is a 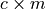 matrix
where
matrix
where  is the number of conservation laws and
is the number of conservation laws and 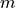 the number of species.
the number of species.
- RoadRunner.getL0Matrix()
Returns the L0 matrix for the current model. The L0 matrix is an (m-r) by r matrix that expresses the dependent reaction rates in terms of the independent rates. m is the number of floating species and r is the rank of the stoichiometry matrix.
- Return type:
numpy.ndarray
- RoadRunner.getLinkMatrix()
Returns the full link matrix, L for the current model. The Link matrix is an m by r matrix where m is the number of floating species and r the rank of the stoichiometric matrix, N.
- Return type:
numpy.ndarray
- RoadRunner.getNrMatrix()
Returns the reduced stoichiometry matrix,
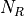 , which will have only r rows where r is the rank of
the full stoichiometry matrix. The matrix will be reordered such that the rows of
, which will have only r rows where r is the rank of
the full stoichiometry matrix. The matrix will be reordered such that the rows of 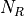 are independent.
are independent.- Return type:
numpy.ndarray
- RoadRunner.getKMatrix()
Returns the K matrix,
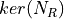 , (right nullspace of Nr)
The K matrix has the structure,
, (right nullspace of Nr)
The K matrix has the structure, ![[I K0]'](../_images/math/adc2c97900ab88829eb36ff8437d7283b0f8a30c.png)
- Return type:
numpy.ndarray
Analysis
- RoadRunner.getFrequencyResponse(startFrequency, numberOfDecades, numberOfPoints, parameterName, variableName, useDB, useHz)
Computes the frequency response. Returns a numpy array with three columns. First column is the frequency, second column the amplitude, and the third column the phase.
- Parameters:
startFrequency – Start frequency for the first column in the output
numberOfDecades (int) – Number of decades for the frequency range, eg 4 means the frequency span 10,000
numberOfPoints (int) – The number of points to generate in the output
parameterName (str) – The parameter where the input frequency is applied, usually a boundary species, eg ‘Xo’
variableName (str) – The amplitude and phase will be output for this variable, usually a floating species, eg ‘S1’
useDB (boolean) – If true use Decibels on the amplitude axis
useHz (boolean) – If true use Hertz on the x axis, the default is rads/sec
For example:
import tellurium as te import roadrunner from matplotlib import pyplot as plt rr = te.loada(""" $Xo -> x1; k1*Xo - k2*x1; x1 -> x2; k2*x1 - k3*x2; x2 ->; k3*x2; k1 = 0.5; k2 = 0.23; k3 = 0.4; Xo = 5; """) rr.steadyState() m = rr.getFrequencyResponse(0.001, 5, 1000, 'Xo', 'x2', True, False) fig = plt.figure(figsize=(10,4)) ax1 = fig.add_subplot (121) ax1.semilogx (m[:,0], m[:,1], color="blue", linewidth="2") ax1.set_title ('Amplitude') plt.xlabel ('Frequency') ax2 = fig.add_subplot (122) ax2.semilogx (m[:,0], m[:,2], color="blue", linewidth="2") ax2.set_title ('Phase') plt.xlabel ('Frequency') plt.show()
- RoadRunner.getRatesOfChange()
Returns the rates of change of all floating species. The order of species is given by the order of Ids returned by getFloatingSpeciesIds()
- Returns:
a named array of floating species rates of change.
- Return type:
numpy.ndarray
>>> rr.getRatesOfChange() MKKK, MKKK_P, MKK, MKK_P, MKK_PP, MAPK, MAPK_P, MAPK_PP [[ 0.000503289, -0.000503289, 0.537508, -0.0994839, -0.438024, 0.061993, 0.108417, -0.17041]]
- RoadRunner.getIndependentRatesOfChange()
Returns the rates of change of all independent floating species. The order of species is given by the order of Ids returned by getIndependentFloatingSpeciesIds()
- Returns:
a named array of independent floating species rates of change.
- Return type:
numpy.ndarray
>>> rr.getIndependentRatesOfChange() MKK_P, MAPK_P, MKKK, MKK, MAPK [[ -0.0994839, 0.108417, 0.000503289, 0.537508, 0.061993]]
- RoadRunner.getDependentRatesOfChange()
Returns the rates of change of all dependent floating species. The order of species is given by the order of Ids returned by getDependentFloatingSpeciesIds()
- Returns:
a named array of dependent floating species rates of change.
- Return type:
numpy.ndarray
>>> rr.getDependentRatesOfChange() MKK_PP, MKKK_P, MAPK_PP [[ -0.438024, -0.000503289, -0.17041]]